Overview
The scripting capabilities of the PIPER application let the user insert custom operation in his workflow by Writing a script. It is also possible to run complete workflows from scripts, this is the so called Batch mode. The scripts are written in Python 2.7, see PIPER Python packages for a reference of the PIPER specific python API.
Running a script
From the Scripting menu any python script can be run at any time. If the script expects parameters, they can be specified in a space separated list. A list of standard script is provided with the application (they are located in share/python/scripting). Additionnaly the user can set a custom script folder from which the python scripts are listed.
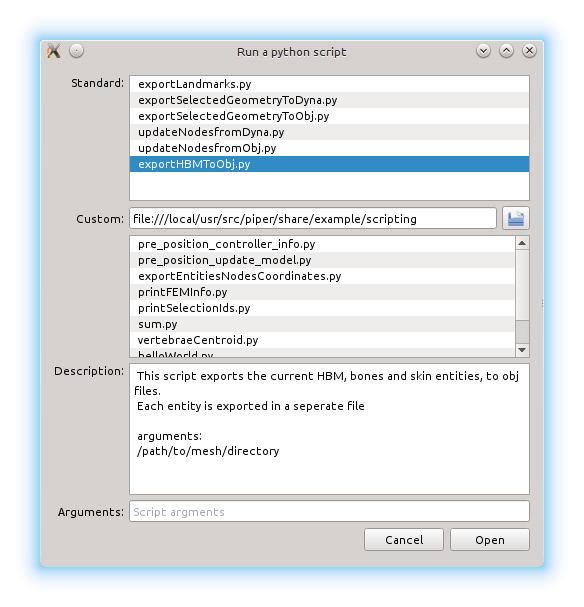
Running a python script with arguments
Writing a script
Using the provided piper python packages, the user can access and modify the current application model, metadata and targets. The following example are available in the share/example/scripting folder.
Basic functionalities
The traditional Hello world ! example (in helloWorld.py):
6 piper.app.logInfo(
"Hello World !")
A script which manipulates its arguments (in sum.py) :
12 piper.app.logInfo(
"a+b = {0}".format(a+b))
14 if (3 != len(sys.argv)):
15 raise RuntimeError(
"Invalid arguments")
17 computeSum(float(sys.argv[1]), float(sys.argv[2]))
Accessing model metadata and geometry
The following example shows how to access the current model and metadata, and also query the anatomy database. The script computes the vertebrae centroids and print the result in the application log (in vertebraeCentroid.py) :
13 model = piper.app.Context.instance().project().model()
17 piper.app.logWarning(
"Model is empty")
20 metadata = piper.app.Context.instance().project().model().metadata()
21 fem = piper.app.Context.instance().project().model().fem()
22 for (name,entity)
in metadata.entities().iteritems() :
24 if piper.anatomyDB.isEntityPartOf(name,
"Vertebral_column",
True):
25 piper.app.logInfo(
"{0}: {1}".format(name, computeCentroid(entity, fem)))
28 def computeCentroid(entity, fem):
29 c = numpy.zeros((3,1))
30 envelopNodesId = entity.getEnvelopNodes(fem)
31 for id
in envelopNodesId :
32 c += fem.getNode(id).get()
33 return (c / len(envelopNodesId)).flatten()
The following example shows how to export the currently selected nodes and elements to an external exportSelectedGeometryToObj.py).
16 def writeObj(fem, nodeIds, elem2DIds, objFilePath):
21 for elemId
in elem2DIds:
22 elem = fem.getElement2D(elemId)
23 if not elem.getType()
in {piper.hbm.ELT_TRI, piper.hbm.ELT_QUAD}:
24 piper.app.logWarning(
"Unsupported: elem: {0} type: {1}".format(elemId, elem.getType()))
30 nodeIdToObjIndex = dict()
31 objIndexToNodeId = dict()
32 with open(objFilePath,
'w')
as objFile:
33 piper.app.logInfo(
"Writting obj file {0} ...".format(objFilePath))
34 objFile.write(
"# File generated by the PIPER application\n")
36 for nodeId
in allNodeIds:
37 nodeIdToObjIndex[nodeId] = currentIndex
38 objIndexToNodeId[currentIndex] = nodeId
39 coord = fem.getNode(nodeId).get().flatten()
40 objFile.write(
"v {0} {1} {2}\n".format(coord[0], coord[1], coord[2]))
42 for elemId
in elem2DIds:
43 elem = fem.getElement2D(elemId)
45 if elem.getType() == piper.hbm.ELT_QUAD:
47 objFile.write(
"f {0} {1} {2}\n".format(nodeIdToObjIndex[idsQuad[0]], nodeIdToObjIndex[idsQuad[1]], nodeIdToObjIndex[idsQuad[2]]))
48 objFile.write(
"f {0} {1} {2}\n".format(nodeIdToObjIndex[idsQuad[0]], nodeIdToObjIndex[idsQuad[2]], nodeIdToObjIndex[idsQuad[3]]))
50 elif elem.getType() == piper.hbm.ELT_TRI:
51 objFile.write(
"f {0} {1} {2}\n".format(nodeIdToObjIndex[ids[0]], nodeIdToObjIndex[ids[1]], nodeIdToObjIndex[ids[2]]))
54 objIdFilePath = objFilePath+
".json"
55 with open(objIdFilePath,
'w')
as objIdFile:
56 piper.app.logInfo(
"Writting obj id file {0} ...".format(objIdFilePath))
57 json.dump(objIndexToNodeId, objIdFile)
59 model = piper.app.Context.instance().project().model()
63 piper.app.logWarning(
"Model is empty")
64 elif 2 != len(sys.argv) :
65 raise RuntimeError(
"Invalid arguments")
67 modelVTK = model.fem().getFEModelVTK()
68 nodeIds = modelVTK.getSelectedNodes()
69 elem2DIds = modelVTK.getSelectedElements(2)
70 piper.app.logInfo(
"nb selected nodes: {0}".format(len(nodeIds)))
71 piper.app.logInfo(
"nb selected elements: {0}".format(len(elem2DIds)))
72 writeObj(model.fem(), nodeIds, elem2DIds, sys.argv[1])
The following example shows how to export entities nodes id in FE code and coordinate (in exportEntitiesNodesCoordinates.py)
14 model = piper.app.Context.instance().project().model()
18 piper.app.logWarning(
"Model is empty")
19 elif not ( len(sys.argv)
in {2,3} ) :
20 raise RuntimeError(
"Invalid arguments")
22 exportDirectory = sys.argv[1]
25 originalHistory =
None
26 if len(sys.argv) == 3:
27 originalHistory = model.history().getCurrent()
28 model.history().setCurrentModel(sys.argv[2])
30 shutil.rmtree(exportDirectory, ignore_errors=
True)
31 os.makedirs(exportDirectory)
32 metadata = model.metadata()
34 piperIdToIdKey = fem.computePiperIdToIdKeyMapNode();
35 for (name,entity)
in metadata.entities().iteritems() :
36 ids = entity.getEntityNodesIds(fem)
37 datFilePath = os.path.join(exportDirectory, name+
".dat")
38 piper.app.logInfo(
"Export {0} nodes from entity {1} to {2}".format(len(ids), name, datFilePath))
39 with open(datFilePath,
'w')
as datFile:
40 datFile.write(
"# Entity {0}\n".format(name))
41 datFile.write(
"# Id x y z\n")
43 coord = fem.getNode(nodeId).get().flatten()
44 datFile.write(
"{0} {1} {2} {3}\n".format(piperIdToIdKey[nodeId].id, coord[0], coord[1], coord[2]))
47 if not originalHistory
is None:
48 model.history().setCurrentModel(originalHistory)
TODO: examples with target creation, TODO: example with data preparation saved to (octave) file, running octave, reading output and preparing targets TODO: doc for piper.app, piper.hbm and piper.anatomydb python packages
Batch mode
The batch mode enable the user to prepare scripts to run at once a sequence of operations. The functions available in batch mode includes Project read/write, import/export, target read/write and modification, module parameters modification and access to PIPER Modules Overview.
The batch mode is started with:
$ piper --batch my_script.py
More options are available, refer to "piper --help" output.
Examples
Target creation
12 leftASISTarget.setName(
"Pelvis_position_1")
13 leftASISTarget.setUnit(piper.hbm.Length_m)
14 leftASISTarget.landmark =
"Left_ASIS"
15 leftASISTarget.setValue({0:0.3, 1:1.2, 2:-0.25})
16 target.add(leftASISTarget)
18 piper.test.expect_eq(piper.hbm.Length_m, leftASISTarget.unit(),
"Error during target creation")
21 hipTarget.setName(
"Hip_position")
22 hipTarget.setUnit(piper.hbm.Length_dm)
23 hipTarget.joint=
"Left_hip_joint"
24 hipTarget.setValue({4:math.radians(45)})
27 piper.test.expect_eq(piper.hbm.Length_dm, hipTarget.unit(),
"Error during target creation")
30 headTarget.setName(
"Head_position")
31 headTarget.frameSource =
"W_Origin"
32 headTarget.frameTarget =
"B_Skull"
33 headTarget.setUnit(piper.hbm.Length_mm)
34 headTarget.setValue({0:200})
35 target.add(headTarget)
37 piper.test.expect_eq(4, target.size(),
"Error during target creation")
Physics based positioning
7 project = piper.app.Project()
10 chilpProjectFile = os.path.join(piper.app.tempDirectoryPath(),
"child.ppj")
12 project.read(chilpProjectFile)
14 project.moduleParameter().setInt(
"PhysPosi",
"autoStopNbIterationMax", 100)
15 project.moduleParameter().setFloat(
"PhysPosi",
"autoStopVelocity", 1e-7)
16 project.moduleParameter().setFloat(
"PhysPosi",
"voxelSize", 0.01)
23 hipTarget.setName(
"Hip_position")
24 hipTarget.joint=
"Left_hip_joint"
25 hipTarget.setValue({4:math.radians(45)})
29 headTarget.setName(
"Head_position")
30 headTarget.setUnit(piper.hbm.Length_mm)
31 headTarget.frameSource =
"W_Origin"
32 headTarget.frameTarget =
"B_Skull"
33 headTarget.setValue({0:200})
34 target.add(headTarget)
36 piper.app.physPosiDeform(project, target)
PIPER Python packages
The PIPER specific functions are made available in batch thanks to several Python packages :


